Modulasi pembungaan untuk efisiensi pemuliaan dan optimalisasi biomassa: Tinjauan molekuler dan bioteknologi
DOI:
https://doi.org/10.70158/buitenzorg.v2i1.18Abstrak
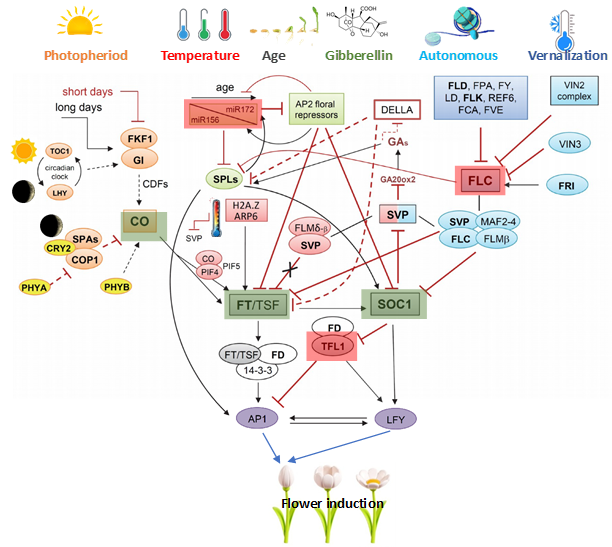
Pengaturan waktu pembungaan merupakan penentu penting keberhasilan reproduksi tanaman dan sifat utama untuk mengoptimalkan adaptasi tanaman, stabilitas hasil, dan efisiensi pemuliaan. Tinjauan ini menyoroti kemajuan terkini dalam jalur molekuler yang mengendalikan pembungaan, termasuk penginderaan fotoperiode, vernalisasi dan respons suhu, pengaturan otonom dan hormonal, dan jaringan integrator bunga. Gen-gen utama seperti FT, SOC1, FLC, TFL1, dan Ghd7 berfungsi sebagai simpul pusat dalam jalur-jalur yang saling berhubungan ini. Penerapan alat rekayasa genetika—termasuk ekspresi gen yang berlebihan, penghapusan yang dimediasi CRISPR/Cas, penyuntingan promotor, dan sistem ekspresi sementara—telah memungkinkan manipulasi fenologi pembungaan yang tepat di berbagai tanaman. Strategi-strategi ini telah mempercepat pemuliaan jalur cepat pada tanaman tahunan beriklim sedang dan tropis dan memfasilitasi peningkatan biomassa vegetatif pada tanaman hijauan dan tanaman industri melalui pembungaan yang tertunda. Namun, penerapan genotipe yang dimodifikasi pembungaan menghadirkan tantangan, termasuk interaksi lingkungan, pertukaran fenologi, pengaturan biosafety, dan dampak ekologis potensial. Arah masa depan harus menekankan integrasi pengendalian waktu pembungaan dengan platform pemuliaan cepat, seleksi genomik, dan desain sifat adaptif iklim, yang disesuaikan dengan kebutuhan khusus spesies dan wilayah. Pendekatan multidisiplin seperti itu akan sangat penting untuk memajukan ketahanan, produktivitas, dan keberlanjutan tanaman dalam kondisi lingkungan yang berubah.
Kata kunci: pengaturan waktu pembungaan, rekayasa genetika, gen FT, pemuliaan cepat, pengoptimalan biomassa
Unduhan
Referensi
Ahmar, S., Zhai, Y., Huibin, H., Yu, K., Khan, M. H. U., Shahid, M., Samad, R. A., Khan, S. U., Amoo, O., Fan, C., & Zhou, Y. (2021). Development of mutants with varying flowering times by targeted editing of multiple SVP gene copies in Brassica napus L. Crop Journal, 10(1), 67–74. https://doi.org/10.1016/j.cj.2021.03.023 DOI: https://doi.org/10.1016/j.cj.2021.03.023
Amasino, R. (2010). Seasonal and developmental timing of flowering. Plant Journal, 61(6), 1001–1013. https://doi.org/10.1111/j.1365313X.2010.04148.x DOI: https://doi.org/10.1111/j.1365-313X.2010.04148.x
Asp, T., Byrne, S., Gundlach, H., Bruggmann, R., Mayer, K. F. X., Andersen, J. R., Xu, M., Greve, M., Lenk, I., & Lübberstedt, T. (2011). Comparative sequence analysis of VRN1 alleles of Lolium perenne with the co-linear regions in barley, wheat, and rice. Molecular Genetics and Genomics, 286(5–6), 433–447. https://doi.org/10.1007/s00438-011-0654-8 DOI: https://doi.org/10.1007/s00438-011-0654-8
Bellec, Y., Guyon-Debast, A., François, T., Gissot, L., Biot, E., Nogué, F., Faure, J. D., & Tepfer, M. (2022). New flowering and architecture traits mediated by multiplex CRISPR-Cas9 gene editing in hexaploid Camelina sativa. Agronomy, 12(8), 1873. https://doi.org/10.3390/agronomy12081873 DOI: https://doi.org/10.3390/agronomy12081873
Boss, P. K., Bastow, R. M., Mylne, J. S., & Dean, C. (2004). Multiple pathways in the decision to flower: enabling, promoting, and resetting. The Plant Cell, 16(suppl_1), S18–S31. https://doi.org/10.1105/tpc.015958 DOI: https://doi.org/10.1105/tpc.015958
Castaings, L., Bergonzi, S., Albani, M. C., Kemi, U., Savolainen, O., & Coupland, G. (2014). Evolutionary conservation of cold-induced antisense RNAs of FLOWERING LOCUS C in Arabidopsis thaliana perennial relatives. Nature Communications, 5, 1–9. https://doi.org/10.1038/ncomms5457 DOI: https://doi.org/10.1038/ncomms5457
Chen, K., Wang, Y., Zhang, R., Zhang, H., & Gao, C. (2019). CRISPR/Cas genome editing and precision plant breeding in agriculture. Annual Review of Plant Biology, 70, 667–697. https://doi.org/10.1146/annurev-arplant-050718-100049 DOI: https://doi.org/10.1146/annurev-arplant-050718-100049
Deng, Q., Wang, Y., Feng, J., Wei, D., Wang, Z. X., & Tang, Q. (2024). Brassica juncea BjuWRKY71-1 accelerates flowering by regulating the expression of SOC1. Sheng Wu Gong Cheng Xue Bao, 40(4), 1017–1028. https://doi.org/10.13345/j.cjb.230400
Deng, W., Ying, H., Helliwell, C. A., Taylor, J. M., Peacock, W. J., & Dennis, E. S. (2011). FLOWERING LOCUS C (FLC) regulates development pathways throughout the life cycle of Arabidopsis. Proceedings of the National Academy of Sciences of the United States of America, 108(16), 6680–6685. https://doi.org/10.1073/ pnas.1103175108 DOI: https://doi.org/10.1073/pnas.1103175108
Deva, C., Dixon, L., Urban, M., Ramirez‐Villegas, J., Droutsas, I., & Challinor, A. (2023). A new framework for predicting and understanding flowering time for crop breeding. Plants, People, Planet, 6(1), 197–209. https://doi.org/10.1002/ppp3.10427 DOI: https://doi.org/10.1002/ppp3.10427
Endo, T., Fujii, H., Omura, M., & Shimada, T. (2020). Fast-track breeding system to introduce CTV resistance of trifoliate orange into citrus germplasm, by integrating early flowering transgenic plants with marker-assisted selection. BMC Plant Biology, 20(1), 1–16. https://doi.org/10.1186/s12870-020-02399-z DOI: https://doi.org/10.1186/s12870-020-02399-z
Flachowsky, H., Roux, P. M. Le, Peil, A., Patocchi, A., Richter, K., & Hanke, M. V. (2011). Application of a high-speed breeding technology to apple (Malus × domestica) based on transgenic early flowering plants and marker-assisted selection. New Phytologist, 192(2), 364–377. https://doi.org/10.1111/j.1469-8137.2011.03813.x DOI: https://doi.org/10.1111/j.1469-8137.2011.03813.x
Fornara, F., de Montaigu, A., & Coupland, G. (2010). SnapShot: control of flowering in Arabidopsis. Cell, 141(3), 3–5. https://doi.org/10.1016/j.cell.2010.04.024 DOI: https://doi.org/10.1016/j.cell.2010.04.024
Gaurha, A., Dewangan, R. K., Minz, V., Shukla, S., Shrivastava, P., & Yamini, Y. (2024). A comprehensive review on fast track breeding of fruit crops: a new approach. Plant Archives, 24(1), 929–937. https://doi.org/10.51470/plantarchives.2024.v24.no.1.127 DOI: https://doi.org/10.51470/PLANTARCHIVES.2024.v24.no.1.127
Ghosh, S., Roy, A., & Dutta, S. (2024). Rapid generation advance methods to fast-track crop breeding: a review. Agricultural Reviews, 45 (4), 693–698. https://doi.org/10.18805/ag.r-2476 DOI: https://doi.org/10.18805/ag.R-2476
Greb, T., Mylne, J. S., Crevillen, P., Geraldo, N., An, H., Gendall, A. R., & Dean, C. (2007). The PHD finger protein VRN5 functions in the epigenetic silencing of Arabidopsis FLC. Current Biology, 17(1), 73–78. https://doi.org/10.1016/j.cub.2006.11.052 DOI: https://doi.org/10.1016/j.cub.2006.11.052
Hanano, S., & Goto, K. (2011). Arabidopsis TERMINAL FLOWER1 is involved in the regulation of flowering time and inflorescence development through transcriptional repression. The Plant Cell, 23(9), 3172–3184. https://doi. org/10.1105/tpc.111.088641 DOI: https://doi.org/10.1105/tpc.111.088641
Hodaei, A., & Werbrouck, S. (2023). Unlocking nature’s clock: CRISPR technology in flowering time engineering. Plants, 12(23), 4020. https://doi.org/10.3390/plants12234020 DOI: https://doi.org/10.3390/plants12234020
Hu, Y., Li, S., & Xing, Y. (2019). Lessons from natural variations: artificially induced heading date variations for improvement of regional adaptation in rice. Theoretical and Applied Genetics, 132(2), 383–394. https://doi.org/10.1007/s00122-018-3225-0 DOI: https://doi.org/10.1007/s00122-018-3225-0
Ionescu, I. A., Møller, B. L., & Sánchez-Pérez, R. (2016). Chemical control of flowering time. Journal of Experimental Botany, 67(1), 27–40. https://doi.org/10.1093/jxb/erw427 DOI: https://doi.org/10.1093/jxb/erw427
Jeong, S. Y., Ahn, H., Ryu, J., Oh, Y., Sivanandhan, G., Won, K. H., Park, Y. D., Kim, J. S., Kim, H., Lim, Y. P., & Kim, S. G. (2019). Generation of early-flowering Chinese cabbage (Brassica rapa spp. pekinensis) through CRISPR/Cas9-mediated genome editing. Plant Biotechnology Reports, 13(5), 491–499. https://doi.org/10.1007/S11816-019-00566-9 DOI: https://doi.org/10.1007/s11816-019-00566-9
Kim, D. H. (2020). Current understanding of flowering pathways in plants: focusing on the vernalization pathway in Arabidopsis and several vegetable crop plants. Horticulture Environment and Biotechnology, 61(2), 209–227. https://doi.org/ 10.1007/s13580-019-00218-5 DOI: https://doi.org/10.1007/s13580-019-00218-5
Komiya, R., Ikegami, A., Tamaki, S., Yokoi, S., & Shimamoto, K. (2008). Hd3a and RFT1 are essential for flowering in rice. Development, 135(4), 767–774. https://doi.org/10.1242/DEV.008631 DOI: https://doi.org/10.1242/dev.008631
Kotoda, N., Hayashi, H., Suzuki, M., Igarashi, M., Hatsuyama, Y., Kidou, S. I., Igasaki, T., Nishiguchi, M., Yano, K., Shimizu, T., Takahashi, S., Iwanami, H., Moriya, S., & Abe, K. (2010). Molecular characterization of FLOWERING LOCUS T-Like genes of apple (Malus × domestica Borkh.). Plant and Cell Physiology, 51(4), 561–575. https://doi.org/10.1093/pcp/pcq021 DOI: https://doi.org/10.1093/pcp/pcq021
Leijten, W., Koes, R., Roobeek, I., & Frugis, G. (2018). Translating flowering time from Arabidopsis thaliana to Brassicaceae and Asteraceae crop species. Plants, 7(4), 111. https://doi.org/10.3390/PLANTS7040111 DOI: https://doi.org/10.3390/plants7040111
Li, X., Shen, C., Chen, R., Sun, B., Li, D., Guo, X., Wu, C., Khan, N., Chen, B., & Yuan, J. (2023). Function of BrSOC1b gene in flowering regulation of Chinese cabbage and its protein interaction. Planta, 258(1), 21. https://doi.org/10.1007/s00425-023-04173-5 DOI: https://doi.org/10.1007/s00425-023-04173-5
Liu, S., Liu, S., Qi, T., Ma, J., Ma, T., Ma, L., & Lin, X. (2016). Ectopic expression of a SOC1 homolog from Phyllostachys violascens alters flowering time and identity of floral organs in Arabidopsis thaliana. Trees-Structure and Function, 30(6), 2203–2215. https://doi.org/10.1007/S00468-016-1445-Y DOI: https://doi.org/10.1007/s00468-016-1445-y
Mallik, M. (2018). Flowering control mechanisms in plants and its importance in crop production and breeding. International Journal of Pure & Applied Bioscience, 6(1), 1033–1038. https://doi.org/10.18782/2320-7051.2796 DOI: https://doi.org/10.18782/2320-7051.2796
Nakashima, N., & Miyazaki, K. (2014). Bacterial cellular engineering by genome editing and gene silencing. International Journal of Molecular Sciences, 15(2), 2773–2793. https://doi.org/10.3390/ijms15022773 DOI: https://doi.org/10.3390/ijms15022773
Odipio, J., Getu, B., Chauhan, R. D., Alicai, T., Bart, R., Nusinow, D. A., & Taylor, N. J. (2020). Transgenic overexpression of endogenous FLOWERING LOCUS T-like gene MeFT1 produces early flowering in cassava. PLoS ONE, 15(3), e0231232. https://doi.org/10.1371/journal.pone.0231232 DOI: https://doi.org/10.1371/journal.pone.0231232
Reed, J. W., Nagatani, A., Elich, T. D., Fagan, M., & Chory, J. (1994). Phytochrome A and phytochrome B have overlapping but distinct functions in Arabidopsis development. Plant Physiology, 104(4), 1139–1149. https://doi.org/10.1104/pp.104.4.1139 DOI: https://doi.org/10.1104/pp.104.4.1139
Sasani, S., Hemming, M. N., Oliver, S. N., Greenup, A., Tavakkol-Afshari, R., Mahfoozi, S., Poustini, K., Sharifi, H. R., Dennis, E. S., Peacock, W. J., & Trevaskis, B. (2009). The influence of vernalization and daylength on expression of flowering-time genes in the shoot apex and leaves of barley (Hordeum vulgare). Journal of Experimental Botany, 60(7), 2169–2178. https://doi.org/10.1093/jxb/erp098 DOI: https://doi.org/10.1093/jxb/erp098
Schiessl, S. (2020). Regulation and subfunctionalization of flowering time genes in the allotetraploid oil crop Brassica napus. Frontiers in Plant Science, 11, 605155. https://doi.org/10.3389/FPLS.2020.605155 DOI: https://doi.org/10.3389/fpls.2020.605155
Shang, L., Tao, J., Song, J., Wang, Y., Zhang, X., Ge, P., Li, F., Dong, H., Gai, W., Grierson, D., Ye, Z., & Zhang, Y. (2023). CRISPR/Cas9-mediated mutations of FANTASTIC FOUR gene family for creating early flowering mutants in tomato. Plant Biotechnology Journal, 22(3), 774–784. https://doi.org/10.1111/pbi.14223 DOI: https://doi.org/10.1111/pbi.14223
Shannon, S., & Meeks-Wagner, D. R. (1991). A mutation in the Arabidopsis TFL1 gene affects inflorescence meristem development. Plant Cell, 3(9), 877–892. https://doi.org/10.2307/3869152 DOI: https://doi.org/10.2307/3869152
Sheldon, C. C., Rouse, D. T., Finnegan, E. J., Peacock, W. J., & Dennis, E. S. (2000). The molecular basis of vernalization: the central role of FLOWERING LOCUS C (FLC). Proceedings of the National Academy of Sciences of the United States of America, 97(6), 3753–3758. https://doi.org/10.1073/pnas.97.7.3753 DOI: https://doi.org/10.1073/pnas.97.7.3753
Soyk, S., Müller, N. A., Park, S. J., Schmalenbach, I., Jiang, K., Hayama, R., Zhang, L., Van Eck, J., Jiménez-Gómez, J. M., Jiménez-Gómez, J. M., & Lippman, Z. B. (2017). Variation in the flowering gene SELF PRUNING 5G promotes day-neutrality and early yield in tomato. Nature Genetics, 49(1), 162–168. https://doi.org/10.1038/NG.3733 DOI: https://doi.org/10.1038/ng.3733
Tao, Z., Shen, L., Liu, C., Liu, L., Yan, Y., & Yu, H. (2012). Genome‐wide identification of SOC1 and SVP targets during the floral transition in Arabidopsis. The Plant Journal, 70(4), 549–561. https://doi.org/10.1111/j.1365-313x.2012.04919.x DOI: https://doi.org/10.1111/j.1365-313X.2012.04919.x
Teklemariam, S. S., Bayissa, K. N., Matros, A., Pillen, K., Ordon, F., & Wehner, G. (2024). Genetic analysis of flowering time of Ethiopian barley accessions under field and climate chamber conditions. Agronomy, 14(12), 3031. https://doi.org/10.3390/agronomy14123031 DOI: https://doi.org/10.3390/agronomy14123031
Tränkner, C., Lehmann, S., Hoenicka, H., Fladung, M., Lenhardt, D., Dunemann, F., Gau, A., Schlangen, K., Malnoy, M., & Flachowsky, H. (2010). Over-expression of an FT-homologous gene of apple induces early flowering in annual and perennial plants. Planta, 232(6), 1309–1324. https://doi.org/10.1007/s00425-010-1254-2 DOI: https://doi.org/10.1007/s00425-010-1254-2
Turck, F., Fornara, F., & Coupland, G. (2008). Regulation and identity of florigen: FLOWERING LOCUS T moves center stage. Annual Review of Plant Biology, 59, 573–594. https://doi.org/10.1146/annurev.arplant.59.032607.092755 DOI: https://doi.org/10.1146/annurev.arplant.59.032607.092755
Wang, F., Li, S., Kong, F., Lin, X., & Lu, S. (2023). Altered regulation of flowering expands growth ranges and maximizes yields in major crops. Frontiers in Plant Science, 14, 1094411. https://doi.org/10.3389/fpls.2023.1094411 DOI: https://doi.org/10.3389/fpls.2023.1094411
Wang, G., Wang, C., Lu, G., Wang, W., Mao, G., Habben, J. E., Song, C., Wang, J., Chen, J., Gao, Y., Liu, J., & Greene, T. W. (2020). Knockouts of a late flowering gene via CRISPR-Cas9 confer early maturity in rice at multiple field locations. Plant Molecular Biology, 104(1–2), 137–150. https://doi.org/10.1007/s11103-020-01031-w DOI: https://doi.org/10.1007/s11103-020-01031-w
Watson, A., Ghosh, S., Williams, M. J., Cuddy, W. S., Simmonds, J., Rey, M. D., Md Hatta, M. A., Hinchliffe, A., Steed, A., Reynolds, D., et al. (2017). Speed breeding: a powerful tool to accelerate crop research and breeding. Nature Plants, 4(1), 23–29. https://doi.org/10.1101/161182 DOI: https://doi.org/10.1038/s41477-017-0083-8
Weng, X., Wang, L., Wang, J., Hu, Y., Du, H., Xu, C., Xing, Y., Li, X., Xiao, J., & Zhang, Q. (2014). Grain number, plant height, and Heading Date7 is a central regulator of growth, development, and stress response. Plant Physiology, 164(2), 735–747. https://doi.org/10.1104/pp.113.231308 DOI: https://doi.org/10.1104/pp.113.231308
Wenzel, S., Flachowsky, H., & Hanke, M.-V. (2013). The Fast-track breeding approach can be improved by heat-induced expression of the FLOWERING LOCUS T genes from poplar (Populus trichocarpa) in apple (Malus × domestica Borkh.). Plant Cell, Tissue and Organ Culture, 115(2), 127–137. https://doi.org/10.1007/s11240-013-0346-7 DOI: https://doi.org/10.1007/s11240-013-0346-7
Xu, F., Rong, X., Huang, X., & Cheng, S. (2012). Recent advances of Flowering Locus T gene in higher plants. International Journal of Molecular Sciences, 13(3), 3773–3781. https://doi.org/10.3390/ijms13033773 DOI: https://doi.org/10.3390/ijms13033773
Yamagishi, N., Sasaki, S., Yamagata, K., Komori, S., Nagase, M., Wada, M., Yamamoto, T., & Yoshikawa, N. (2011). Promotion of flowering and reduction of a generation time in apple seedlings by ectopical expression of the Arabidopsis thaliana FT gene using the apple latent spherical virus vector. Plant Molecular Biology, 75(1), 193–204. https://doi.org/10.1007/s11103-010-9718-0 DOI: https://doi.org/10.1007/s11103-010-9718-0
Zhang, B., Wang, L., Zeng, L., Zhang, C., & Ma, H. (2015). Arabidopsis TOE proteins convey a photoperiodic signal to antagonize CONSTANS and regulate flowering time. Genes and Development, 29(9), 975–987. https://doi.org/10.1101/gad.251520.114 DOI: https://doi.org/10.1101/gad.251520.114
Zhao, J., Huang, X., Ouyang, X., Chen, W., Du, A., Zhu, L., Wang, S., Deng, X. W., & Li, S. (2012). OsELF3-1, an ortholog of Arabidopsis EARLY FLOWERING 3, regulates rice circadian rhythm and photoperiodic flowering. PLoS ONE, 7(8), e43705. https://doi.org/10.1371/journal.pone.0043705 DOI: https://doi.org/10.1371/journal.pone.0043705
Zhao, Z., Yu, Y., Meyer, D., Wu, C., & Shen, W. H. (2005). Prevention of early flowering by expression of FLOWERING LOCUS C requires methylation of histone H3 K36. Nature Cell Biology, 7(12), 1156–1160. https://doi.org/10.1038/ncb1329 DOI: https://doi.org/10.1038/ncb1329
Zhou, S., Zhu, S., Cui, S., Hou, H., Wu, H., Hao, B., Cai, L., Xu, Z., Liu, L., Jiang, L., Wang, H., & Wan, J. (2021). Transcriptional and post‐transcriptional regulation of heading date in rice. New Phytologist, 230(3), 943–956). https://doi.org/10.1111/nph.17158 DOI: https://doi.org/10.1111/nph.17158
Zhu, C., Zheng, X., Huang, Y., Ye, J., Chen, P., Zhang, C., Zhao, F., Xie, Z., Zhang, S., Wang, N., Li, H., Wang, L., Tang, X., Chai, L., Xu, Q., & Deng, X. (2019). Genome sequencing and CRISPR/Cas9 gene editing of an early flowering mini-citrus (Fortunella hindsii). Plant Biotechnology Journal, 17(11), 2199–2210. https://doi.org/10.1111/PBI.13132 DOI: https://doi.org/10.1111/pbi.13132
Unduhan
Diterbitkan
Cara Mengutip
Terbitan
Bagian
Lisensi
Hak Cipta (c) 2025 Buitenzorg: Journal of Tropical Science

Artikel ini berlisensiCreative Commons Attribution-ShareAlike 4.0 International License.
The article is licensed under a Creative Commons Attribution-ShareAlike 4.0 International License (CC BY-SA), which allows both Authors and Readers to copy and distribute the material in any format or medium, as well as modify and create derivative works from it for any purpose, provided that appropriate credit is given (by citing the article or content), a link to the license is provided, and it is indicated if any changes were made. If the material is modified or used to create derivative works, the contributions must be distributed under the same license as the original.







