Micromorphological Characterization and DNA Barcoding of Durio macrantha Kosterm. from the Bogor Botanical Garden Collection
DOI:
https://doi.org/10.70158/buitenzorg.v2i1.16Abstract
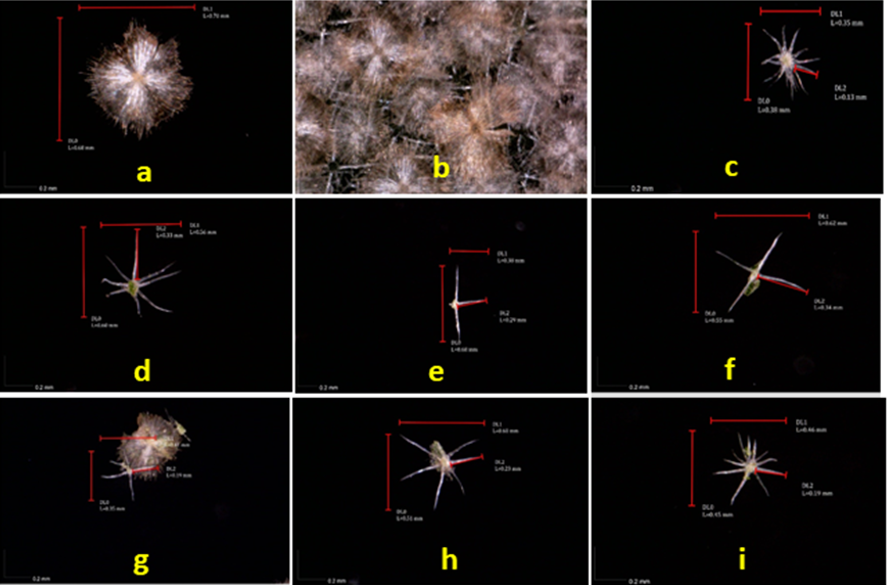
Durio macrantha is an endemic species first discovered in 1981 in Gunung Leuser National Park, North Sumatra, and has been conserved at the Bogor Botanical Garden since 1994. This study aims to characterize the micromorphology and DNA profile D. macrantha using DNA barcoding with rbcL and ITS gene markers. Sample of D. macrantha were collected from the Bogor Botanical Gardens for analysis. Micromorphological analysis revealed that the leaves exhibit three vein types (primary, secondary, and tertiary) with a pentagonal venation pattern and a tri-veinlet veinlet pattern. Additionally, eight types of trichomes were observed on the abaxial surface. Molecular characterization showed that the rbcL sequence was 563 bp long, with nucleotide composition T (27.9%), C (21.7%), A (27.9%), and G (22.6%). The ITS sequence was 960 bp long, with nucleotide composition T (15.4%), C (33.4%), A (19.0%), and G (32.2%). BLAST analysis of both rbcL and ITS genes revealed a high level of similarity between D. macrantha and D. zibethinus. This study provides fundamental data supporting the conservation and further research of D. macrantha, particularly in morphological and molecular aspects.
Downloads
References
Alcántara-Ayala, O., Oyama, K., Ríos-Muñoz, C. A., Rivas, G., Ramirez-Barahona, S., & Luna-Vega, I. (2020). Morphological variation of leaf traits in the Ternstroemia lineata species complex (Ericales: Penthaphylacaceae) in response to geographic and climatic variation. PeerJ, 8, e8307. https://doi.org/10.7717/peerj.8307 DOI: https://doi.org/10.7717/peerj.8307
Aprilianingsih, R., Baiq, F. W., & Hariri, M. R. (2022). DNA barcode of Homalomena pexa inferred from internal transcribed spacer region. Jurnal Riset Biologi dan Aplikasinya, 4(2), 69–74. https://doi.org/10.26740/jrba DOI: https://doi.org/10.26740/jrba.v4n2.p69-74
Aprilianti, P. (2019). Konservasi ex-situ Durio spp. di Kebun Raya Bogor (Jawa Barat) dan Kebun Raya Katingan (Kalimantan Tengah). Prosiding Seminar Nasional Masyarakat Biodiversitas Indonesia, 5, 123–128. https://doi.org/10.13057/psnmbi/m050123
Awaliah, S. R., & Polosoro, A. (2024). A comparative study of DNA barcoding markers for bamboo. Buitenzorg: Journal of Tropical Science, 1(2), 30–41. https://doi.org/10.70158/buitenzorg.v1i2.9 DOI: https://doi.org/10.70158/buitenzorg.v1i2.9
Bafeel, S. O., Arif, I. A., Bakir, M. A., Al Homaidan, A. A., Al Farhan, A. H., & Khan, H. A. (2012). DNA barcoding of arid wild plants using rbcL gene sequences. Genetics and Molecular Research, 11(3), 1934–1941. https://doi.org/10.4238/2012.July.19.12 DOI: https://doi.org/10.4238/2012.July.19.12
Barthlott, W., & Neinhuis, C. (1997). Purity of the sacred lotus, or escape from contamination in biological surfaces. Planta, 202, 1–8. https://doi.org/10.1007/s004250050096 DOI: https://doi.org/10.1007/s004250050096
Basith, A. (2015). Peluang gen rbcL sebagai DNA barcode berbasis DNA kloroplas untuk mengungkap keanekaragaman genetik padi beras hitam (Oryza sativa L.) lokal Indonesia. Seminar Nasional XII Pendidikan Biologi FKIP UNS, 938–941.
Berry, V., & Gascuel, O. (1996). On the interpretation of bootstrap trees: Appropriate threshold of clade selection and induced gain. Molecular Biology and Evolution, 13(7), 999–1011. https://doi.org/10.1093/molbev/13.7.999 DOI: https://doi.org/10.1093/molbev/13.7.999
Cheng, T., Xu, C., Lei, L., Li, C., Zhang, Y., & Zhou, S. (2016). Barcoding the kingdom Plantae: new PCR primers for ITS regions of plants with improved universality and specificity. Molecular Ecology Resources, 16(1), 138–149. https://doi.org/10.1111/1755-0998.12438 DOI: https://doi.org/10.1111/1755-0998.12438
Corpuz, T. A., Songpol, S., Salma, I., & Bhag, M. (2007). Descriptors for durian (Durio zibethinus Murr.). In Descriptors for durian (Durio zibethinus Murr.). Bioversity International.
Edgar, R. C. (2004). MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics, 5, 1–19. https://doi.org/10.1186/1471-2105-5-113 DOI: https://doi.org/10.1186/1471-2105-5-113
Ekanayaka, A. H., Karunarathna, S. C., Tibpromma, S., Dutta, A. K., Tennakoon, D. S., Karunarathna, A., Chukeatirote, E., Dai, D. Q., Stephenson, S. L., Maharachchikumbura, S. S. N., Liu, C., & Phillips, A. J. L. (2025). Species evolution: cryptic species and phenotypic noise with a particular focus on fungal systematics. Frontiers in Cellular and Infection Microbiology, 15. https://doi.org/10.3389/fcimb.2025.1497085 DOI: https://doi.org/10.3389/fcimb.2025.1497085
Ellis, B., Douglas, C. D., Leo, J. H., Kirk, R. J., John, D. M., Peter, W., & Scott, L. W. (2009). Manual of leaf architecture. Systematic Botany, 34(4), 825. https://doi.org/10.1600/036364409790139682 DOI: https://doi.org/10.1600/036364409790139682
Felsenstein, J. (1985). Confidence limits on phylogenies: an approach using the bootstrap. Evolution, 39(4), 783–791. https://doi.org/10.1111/j.1558-5646.1985.tb00420.x DOI: https://doi.org/10.1111/j.1558-5646.1985.tb00420.x
Fujita, H., & Mochizuki, A. (2006). The origin of the diversity of leaf venation pattern. Developmental Dynamics, 235(10), 2710–2721. https://doi.org/10.1002/dvdy.20908 DOI: https://doi.org/10.1002/dvdy.20908
GBIF Secretariat (2025). GBIF backbone taxonomy. Retrieved from https://www.gbif.org/species/4073099
Gielly, L., & Taberlet, P. (1994). The use of chloroplast DNA to resolve plant phylogenies: noncoding versus rbcL sequences. Molecular Biology and Evolution, 11(5), 769–777. https://doi.org/10.1093/oxfordjournals.molbev.a040157 DOI: https://doi.org/10.1093/oxfordjournals.molbev.a040157
Hickey, L. J. (1973). Classification of the architecture of Dicotyledonous leaves. American Journal of Botany, 60(1), 17–33. https://doi.org/10.2307/2441319 DOI: https://doi.org/10.1002/j.1537-2197.1973.tb10192.x
Hollingsworth, P. M., Graham, S. W., & Little, D. P. (2011). Choosing and using a plant DNA Barcode. PLoS ONE, 6(5), 1–13. https://doi.org/10.1371/journal.pone.0019254 DOI: https://doi.org/10.1371/journal.pone.0019254
Kostermans, A. J. G. H. (1992). An important economical new Durio species from Northern Sumatra. Economic Botany, 46(3), 338–340. http://www.jstor.org/stable/4255452
Kress, W. J., & Erickson, D. L. (2007). A two-locus global DNA barcode for land plants: the coding rbcL gene complements the non-coding trnH-psbA spacer region. PLoS ONE, 2(6), e508. https://doi.org/10.1371/journal.pone.0000508 DOI: https://doi.org/10.1371/journal.pone.0000508
Magandhi, M., Sobir, Wahyu, Y., Sudarmono, & Matra, D. D. (2024). DNA barcoding of endemic durian kura-kura in West Kalimantan Indonesia. Sabrao Journal of Breeding and Genetics, 56(5), 1799–1810. https://doi.org/10.54910/sabrao2024.56.5.5 DOI: https://doi.org/10.54910/sabrao2024.56.5.5
Nurhasanah, Sundari, & Papuangan, N. (2019). Amplification and analysis of rbcL gene (ribulose-1,5-bisphosphate carboxylase) of clove in Ternate Island. IOP Conference Series: Earth and Environmental Science, 276, 012061. https://doi.org/10.1088/1755-1315/276/1/012061 DOI: https://doi.org/10.1088/1755-1315/276/1/012061
POWO (2025). Plants of the world online. The Royal Botanic Gardens, Kew. Retrieved from https://powo. science.kew.org/taxonurn:lsid:ipni.org:names:960814-1
Priyanti, P., Chikmawati, T., Sobir, S., & Hartana, A. (2015). Leaf trichome morphology of Durio kutejensis landraces from Kalimantan. Makara Journal of Science, 19(1), 37–42. https://doi.org/10.7454/mss.v19i1.4588 DOI: https://doi.org/10.7454/mss.v19i1.4588
Purty, R., & Chatterjee, S. (2016). DNA barcoding: an effective technique in molecular taxonomy. Austin Journal of Biotechnology & Bioengineering, 3(1), 1059.
Rahman, W. (2021). Durio macrantha. The IUCN Red List of Threatened Species 2021: e.T139572994A139572997. Retrieved from https://dx.doi.org/10.2305/IUCN.UK.2021-2.RLTS.T139572994A139572997.en DOI: https://doi.org/10.2305/IUCN.UK.2021-2.RLTS.T139572994A139572997.en
Salma, I. (1999). The taxonomic significance of trichome morphology in the genus Durio (Bombaceae). Garden’s Bulletin Singapore, 51, 55–70. https://biostor.org/reference/140163
Shehata, F. A., Hamdy, R., & Hafez, R. M. (2024). Biosystematic studies of genus Withania Pauquy in Egypt. Scientific Reports, 14, 21754. https://doi.org/10.1038/s41598-024-71500-5 DOI: https://doi.org/10.1038/s41598-024-71500-5
Tamura, K., Stecher, G., & Kumar, S. (2021). MEGA11: molecular evolutionary genetics analysis version 11. Molecular Biology and Evolution, 38(7), 3022–3027. https://doi.org/10.1093/molbev/msab120 DOI: https://doi.org/10.1093/molbev/msab120
Thompson, J. D, Higgins, D. G., & Gibson, T. J. (1994). CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Research, 22(22), 4673–4680. https://doi.org/10.1093/nar/22.22.4673 DOI: https://doi.org/10.1093/nar/22.22.4673
Tozin, L. R. D. S., Stefany, C. D. M. S., & Tatiane, M. R. (2016). Non-glandular trichomes in Lamiaceae and Verbenaceae species: morphological and histochemical features indicate more than physical protection. New Zealand Journal of Botany, 54(4), 446–457. https://doi.org/10.1080/0028825X.2016.1205107 DOI: https://doi.org/10.1080/0028825X.2016.1205107
Werker, E. (2000). Trichome diversity and development. Advances in Botanical Research, 31, 1–35. https://doi.org/10.1016/0065-2296(00)31005-9 DOI: https://doi.org/10.1016/S0065-2296(00)31005-9
Downloads
Published
How to Cite
Issue
Section
License
Copyright (c) 2025 Buitenzorg: Journal of Tropical Science

This work is licensed under a Creative Commons Attribution-ShareAlike 4.0 International License.
The article is licensed under a Creative Commons Attribution-ShareAlike 4.0 International License (CC BY-SA), which allows both Authors and Readers to copy and distribute the material in any format or medium, as well as modify and create derivative works from it for any purpose, provided that appropriate credit is given (by citing the article or content), a link to the license is provided, and it is indicated if any changes were made. If the material is modified or used to create derivative works, the contributions must be distributed under the same license as the original.






